Computer-Human Virus. Is it possible?
March 23rd, 2021 | by Saren Tasciyan | posted in Biology, Data, Hacking, Science, Software

I know how it sounds… Like “I am really hard” to make some story. Of course virus and computer viruses, similar names, why not bring them together? But these are from 2 different worlds! What do they have in common, except for the similarity of self preservation. Let me clear something, this is not science but more science fiction. But I try to stay within the boundaries of current knowledge and try to portray a possibility. A possibility, which is alone so interesting, that can’t be left unmarked. Can there be a human-computer or computer-human virus? What I mean with that is not some sort of Android virus (pun intended). What I mean is an infectious agent, which can jump between humans and computers, while infecting both of them and spreading also between computers and humans. Even though highly highly highly unlikely, I think it is technically possible and that I find interesting. So let’s dive into the biology, computers, data and cybersecurity!
Disclaimer: I am not a cybersecurity expert. However, I love to learn about it. I have written a few computer viruses and hacked into a few computers in the past. But I am definitely not an expert. I have a biology background and a small business in IT. Please keep in mind that this article was written as quickly as possible. If you find factual or other types of mistakes, please contact me. I will correct them. Also please let me know if you find something not so clear.
Especially in 2021 we all know viruses spread from host to host and take over host cells to produce itself. Often, this results in a disease but not all viruses actually cause symptoms. Some silently survive in the host for long times (e.g. herpes, HPV etc.). We all know also that viruses can spread from one species into another. Once in the host cell, they give instructions to the host cell machinery to copy and release itself. Similarly, computer viruses spread via network or infected devices. They insert instructions to either into existing applications or the host operating system. Unintentionally, host computer runs these instructions on the CPU and spreads the virus further to other systems. So far so good and similar. But how can these viruses jump between biology and computers?

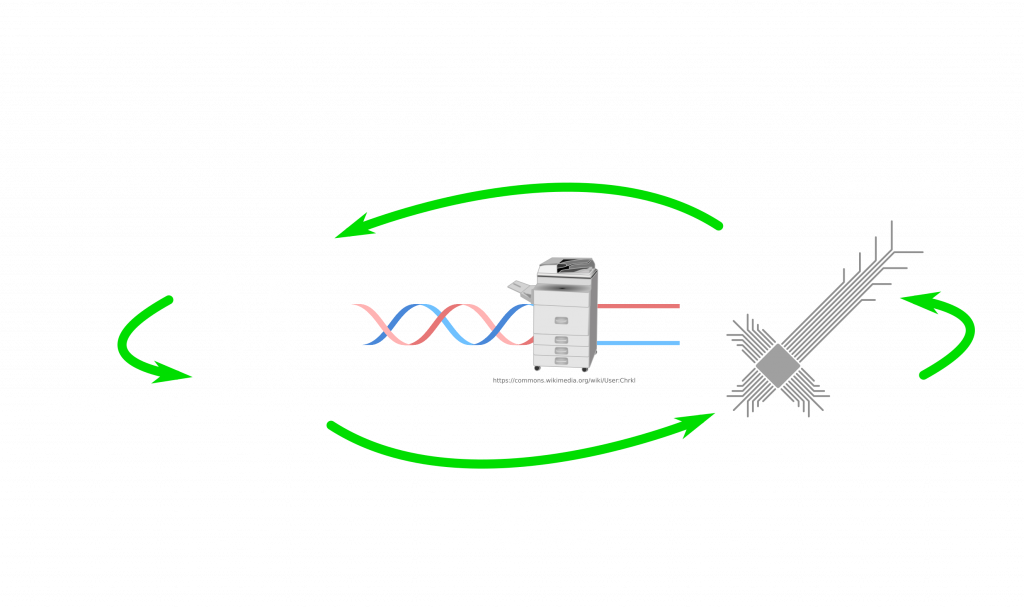
Let’s imagine that any human computer virus can at least propagate either between humans or between computers. This is something that biological and computer viruses do already. Nothing surprising about that. We will call both sides domains. So we have a biological and a digital domain.

First fundamental problem is information. Biology is based on genetic materials, which can be DNA (or RNA). Both of these molecules are similar but they are chemicals after all. Computer software lives on data, which can be stored in different ways but eventually it is an abstract concept. It can be on a magnetic disk, volatile on a transistor network (e.g. RAM), on an optical disk as bunch of holes. Furthermore, smallest genetic information unit is in 2 bits. You can store 4 different cases in 2 bits. This corresponds to A T (or U in RNA) C G, bases in DNA. Software is stored in single bits, hence 2 states: 0 or 1. DNA can be considered as 00, 01, 10, 11. Maybe you have heard of bytes (8 bits). 4 bits are called a nibble. 2 bits are balled a crumb or dibit. This means, by having a lookup table, one can assign bits to nucleic acid sequence in a DNA. This gave the idea for researchers to store data in DNA. Nice thing about this is that DNA is quite stable and one can multiply it in large numbers thanks to the tools provided by life itself. Researchers already managed this. Question is of course, once you store it, how do you read it? Well, you may have heard of sequencing new COVID variants. Sequencing technology does exactly this this. It converts chemically stored ordered bases, which is the data, into digitally stored bits. This enables storage of huge amounts of data in an extremely small space for over large amount of time. You probably, won’t have it in your device to store your pictures, because it is still a tedious process. However, this technology can enable us to compactly store data for the future, in case someone might need it… But I often repeat myself that biology is messy. Hence, you need some extra care how you are storing data as it is open for errors. Nevertheless, there are ways to circumvent this issue. So we can accept that there can be conversion between DNA stored data and computer stored data.
What is software? In essence software itself is data as well. A special kind of data, which contains instructions for a CPU to perform tasks. These instructions are automatically executed thanks to circuitry in a CPU. Those instructions, once engineered carefully, can either create your favorite app, your e-banking website or a computer virus. These instructions are small amounts of data organized in a larger application. These instructions exist in the digital domain. Similarly, life has evolved to process instructions. These instructions as we know it today are copies of the original genetic material in a slightly different chemical form: RNA. More precisely mRNA. Like a processor, ribosomes of the cell translates these into proteins. Proteins then fulfill certain functions. Even though this is an over(over(oversimplification)) but it is enough for the purpose of this article.
First question would be, can you store information on a DNA level to contain design instructions of itself? Well scientists achieved this as well. They stored DNA in free form (cell-free DNA storage) in a 3D printed object. If someone would have found this somewhere and got the crazy idea to sequence a 3D printed object (why not? right?), that person could find the model of the object within the sequence. Since software itself is data, one can store that or any kind of instructions set in a DNA sequence as well. So far, everything I discussed has already been done… From now on, this article is a thought experiment.
So how can we connect both sides in that picture? Let’s place the essential components first! From DNA to digital we need sequencers. How about from digital to DNA? If I have a sequence on my computer, how do I get it as a real DNA molecule? Well, for that there are synthesizers. Machines, which make strands of DNA based on the given sequence. The problem with both directions is that our machines are terrible at making very long strands or even sequence them. Often we chop it up and read/write chunks, which we can then glue/merge with others. But so far so good. We have a bridge between DNA and digital data.
Now consider the easiest but also most terrifying path! Imagine we live in the year 2102 and people receive gene therapies. We inhale an aerosol cloud to stay young or something. Doesn’t matter what for. The cloud contains a gene delivery method. These genes are automatically synthesized in machines connected to network. A well designed computer virus may change the sequence and fool the system. Instead of the real therapy, a long eradicated virus sequence (e.g. small pox) is synthesized. Since, it is eradicated, no human receives vaccination anymore and you have a computer virus mediated viral infection!
If we consider the other way around, I will argue that this is a more challenging path. Let’s say a viral strain is naturally circulating around. In the year, 2102 we sequence a lot. Some of the sequencers receive samples with the viral DNA. By default, like our images, songs, movies on our computers, such data isn’t expected to contain instructions for the CPU. Therefore, it won’t be executed. For instance, an .exe file on Windows is executable and contains instructions. If you change that extension to .jpg, your operating system will start to think that it is a picture file now. Double clicking that file will show you a confused image application. The contents aren’t JPEG. Computers handle different types of data differently. Our hypothetical virus needs to somehow execute instructions on the host computer. But how?
I need to mention field of bioinformatics here. The genetic code sequenced doesn’t tell us much. We need to interpret the genetic code somehow. The data itself will look something like this:
...ATGTCAAGCTCTTCCTGGCTCCTTCTCAGCCTTGTTGCTGTAACTGCTGCTCAGTCCACCATTGAGGAACA...If you look at this code you probably won’t understand anything. That’s now because you aren’t bioinformatician. That’s because we humans can’t easily deal with this kind of data. Probably, in the past people decoded genes by hand. Now we are using computer algorithms to annotate sequences to biological units/meanings. The code is by the way human ACE2 gene (protein, which sars-cov-2 binds) proteins coding sequence starting from the start codon.
Such algorithms find patterns in sequences to identify parts with special meanings. Most simply example could be the start codon: ATG. This marks the first aminoacid to be translated from mRNA, a methionine (see -M- in the above picture). But it is often more complicated than finding any ATG. Not every ATG is a start codon. There are other factors like promoter regions etc. Many sequences, which are recognized by proteins don’t need to be exactly one sequence. Small or big differences may still be recognized, which plays a huge role in turning on/off genes but also how much a gene is turned on. Solving these problems can be a difficult task and also still being researched extensively. If you are a programmer, you may already be thinking about regular expressions. Those would be alone a nightmare to identify such sequences.
Here comes the cyber security part. But we should first talk about computer programs. Computers use memory for quick access to data during run time (e.g. RAM). RAM is orders of magnitude faster than reading from hard drives or SSDs. But to gain even more speed, CPUs have their own small but even faster memory, which is called cache. Current CPUs have complex algorithms to fetch more data from RAM simultaneously into cache to reduce this process. If by chance next step requires something in the cache, then CPU can directly access it without the need of RAM access. Normally, operating system (OS) has the full power over all these resources. Each process still needs to manage own resources. Often the processes behave as if all the resources are there for them. They only wait, if they need further input or event. During this time they sleep. But many processes want to do many things at the same time. OS then provides resources and CPU time to processes as fairly as possible. Sometimes, it pauses one process and starts another. We users don’t notice this as the OS can switch rapidly between processes. And nowadays almost all the CPUs in notebooks and phones have multiple cores to host several processes simultaneously. But the crucial bit if information here is that each process has to demand memory from the OS, when they need it. That memory can be granted by the OS. After this point that process owns that memory. No other process (unless with special rights) is allowed to access that memory. This is for safety. It would be terrible if your application “X” could access the application “Y” memory and steal your passwords. Also even more catastrophic, a malicious process may access OS resources (OS has its own memory) and either read or manipulate. This can lead to an elevated access into the system. Computer malwares (or viruses) try to achieve that. There are some unforeseen bugs, security holes in either OS, device drivers or even badly designed features, which let’s computer viruses access otherwise restricted resources. Computer hackers try to find and identify such vulnerabilities.
On a side note, vulnerabilities can occur on many levels. From large organizations to individuals, applications, hardware drivers, OS and even the hardware itself.
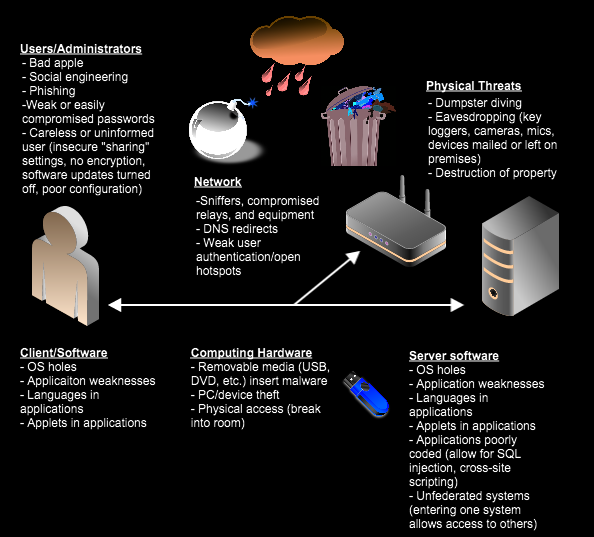
Why did I talk about all that? Because we need to understand how a computer virus can spread and how feasible is it? A “nicely” engineered computer virus can cause a lot of damage. This is a growing concern in many industries, including healthcare, energy and finance. Imagine, half of a country’s power stations shut down and it takes weeks to identify the problem or a hospital system collapsing due to a computer malfunction. Cybersecurity is a real threat and needs to be taken very seriously. Here are some famous examples:
Stuxnet causes substantial damage to Iranian nuclear program
Major hack on US Federal Government (2020)
A computer worm that can slow down the internet
Back to the topic… How can a DNA sequence data on a computer can even manage all that? It isn’t even treated as an executable file. It won’t get any chance to run any instructions on the CPU. Right? Do you remember that each process manages their own memory and the OS doesn’t let processes to access memory other than theirs? But we also know that the DNA sequence is treated as normal data to be analyzed and annotated. Is it possible that a bug or a mistake in the analysis code can result in misallocation of the sequence data into an executable part of the memory? Even more so to run it on a system level? Hmm. It would have been nice to have a real life example… Oh wait, I remember something: stagefright. This was a bug in Android (what a coincidence 🙂 ), which let attackers to run operations and gain privileged access by sending a specially crafted MMS message. This was a zero day vulnerability, meaning that nobody knew this vulnerability existed. This puts systems into a particular danger. Sometimes, people know and choose not to patch. Most importantly, this required no user interaction. Often hackers rely on uninformed socially engineered user interactions. Not in this case. The mechanism behind this vulnerability was an integer overflow bug in a library written in C++. Languages like C or C++ are great for performance but it puts the burden of memory management on the programmer. There can be cases, where the programmer misses a certain case, where a memory overflow or leak can happen. This can result in unexpected results. In this case, one can corrupt the heap memory, which is a freely and dynamically allocated memory.These kind of attacks are also not limited to OS. Here is an Apple version. Anyhow, most important lesson from this to learn is that an innocent piece of data can become a malicious instruction set. There are several types of software vulnerabilities. Some target mistakes in memory management, others input check (malformed inputs can cause troubles) or even concurrency (running multiple instructions/threads simultaneously).
Another more unlikely attack surface could be a code injection attack. This is actually highly unlikely, as I am not aware of SQL (a database query language). Also the way DNA sequence is stored, wouldn’t provide much of an injection opportunity. These kind of attacks often appear in queries, where a request is made in a string format. Sequence in a string format would only include letter A, T, C and G. You can’t write much of code with that. A binary interpretation, may allow though. In this case 4 letters together would form a byte. This allows any character to be written with multiple nucleotides. However, binary data is also handled differently and mostly in the form of hexadecimal representation. We use the decimal system, meaning we use the base 10 to represent numbers. We have 10 characters (0, 1 …. 8, 9). If we want to go higher than 9, we start using a second character e.g. 9 + 1 = 10. Hexadecimal (hex) system uses 16 characters: 0, 1, …. 8, 9, A, B, C, D, E, F. For instance, A means 10, F means 15. F + A = 19 (in hex or 25 in decimal). To avoid confusion in computer code we give hex numbers like this: 0x19. This prevents confusion both for the humans also for the code. A hex byte array can contain a malicious code but it is difficult to trick the system to run it. Overall an injection attack is where a request is made to server, where instead of a simple string a carefully crafted command is inserted to confuse the server. e.g.
SELECT * FROM Users WHERE UserId = userid (normal request)
SELECT * FROM Users WHERE UserId = userid OR 1=1 (simple SQL injection)
1=1 returns always true so whole users table would be returned instead of one. It wasn’t meant to access this data. There are also web based vulnerabilities, which are less relevant for our topic. Therefore, memory leaks or memory attacks seem to be better suited for this purpose.
I think you may guess already, what I mean. Theoretically, a DNA sequence may exploit a similar vulnerability in a DNA sequence analyzer software and spread over network. If this virus spreads between computers, it may eventually end up in a DNA synthesizer. If it is able to recognize that, it may convert itself back into DNA. This is a long stretch, obviously. But for this article we care about the possibility.
Once instructions can be run on the system, virus may try to gain a privilege escalation. Processes run with restricted access to system resources due to security reasons. You don’t want websites you are visiting to gain access to your files and your operating system (OS). Therefore, the javascript code the website is running on your computer is first restricted by your browser, second your browser is restricted by your OS. An attacker, who can at least run javascript on your browser, would want to escape this “prison” in your system. This is called privilege escalation. This can be done in different ways and there has been many examples in the past. Hence, this is not a new concept.
Edit: After finishing the article I came across this: It turns out to be something similar was done in 2017. Researchers tried to use exploits to infect sequence analyzer software! In this case the simple exploit was extremely short. However, this probably would only succeed in one-way transmission.
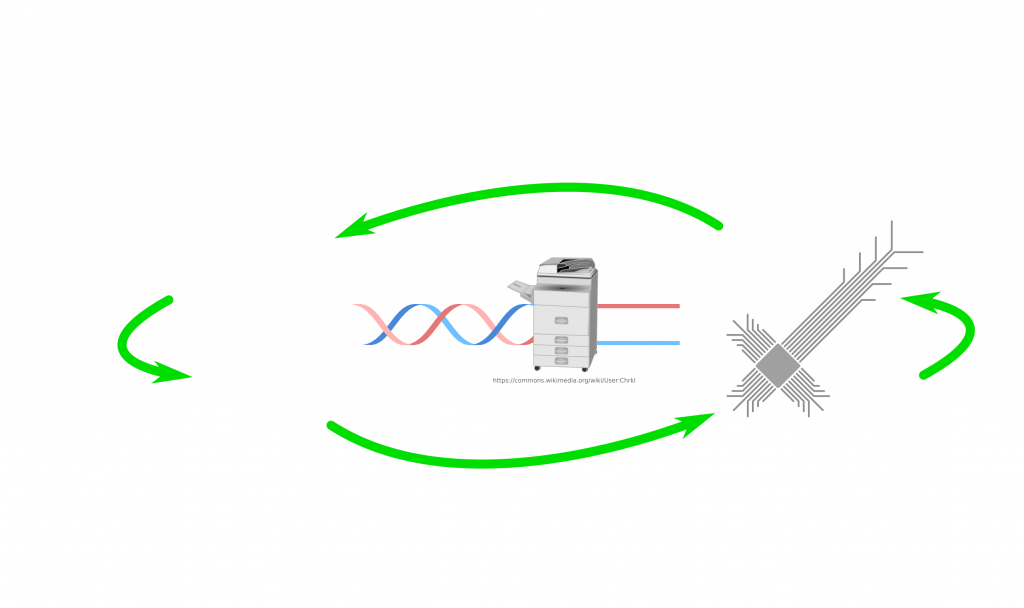
How about biology? Sure you have the DNA but can it infect? Maybe in the form of RNA. Not all viruses use DNA as genetic material to spread. There are many RNA viruses. When it comes to RNA viruses, we need to talk about positive and negative sense RNA. The central dogma in molecular biology states (by Francis Crick) that DNA is transcribed into RNA and RNA is translated into protein. We know that this is generally true but there are exceptions. Some RNA viruses can either replicate directly their RNA and some need to synthesize DNA and then make multiple RNA copies of that DNA. They eventually need to have an RNA molecule to make the viral proteins. A virus carries positive sense RNA, if that RNA molecule is ready for translation (protein making). In contrast, the negative sense needs to be used to make positive sense RNA. For that operation, viral proteins are required. Viruses pack these proteins with their negative sense RNA. Once the virus enters the cell, positive sense RNA generation can start. So, a positive sense RNA doesn’t require anything. Even a purified form can be infectious. Once it enters a cell, immediately cell can translate viral proteins. Those proteins can replicate viral RNA and viral proteins, which leave the cell the infect others. Maybe, for our hypothetical virus, RNA is a better solution to spread from machines to humans. Of course, there also RNA synthesizers.
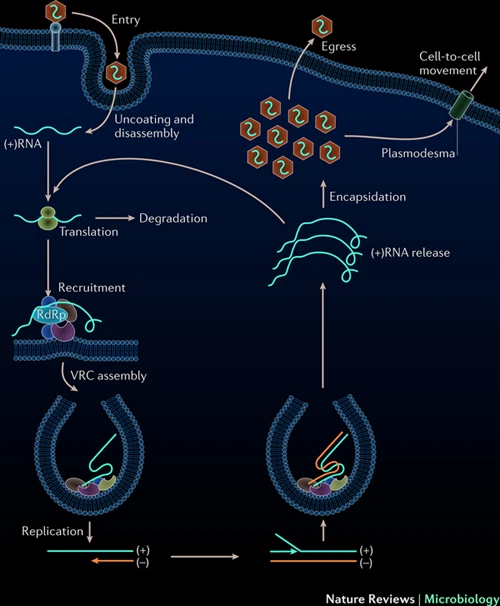
We have established possible routes for human to machine and machine to human infection. Now the question is, how can such a virus be encoded? First problem is to carry all the information within one data strip. The nucleic acid sequence must contain information for both domains (human and digital). This is very tricky, because viruses are restricted in their genetic material size. Viruses are in constant battle between their needs (proteins) and genome size. A genome, which is too large will make them easier to detect by the host systems and also it will be a replication challenge. This means that viruses need to pack most of their genes in a tiny genome. So much so that many genes actually overlap. Sometimes, proteins are created together and the virus relies on host enzymes, which cut them into multiple proteins. Now it needs also carry all that digital instruction set. Imagine the software part to be 50 kilobytes, which is equal to 50000 bytes. This is 400000 bits of data. In terms of sequence length, this would take up at least 200000bp (nucleotides). SARS-CoV-2 is 29903bp long. 6-7x extra material! Even if it would fully overlap with the genes, there is still a large extra bit of information. To my knowledge largest human viruses are herpes viruses, some up to ~235000bp. This is still too small. And this without considering fail safe methods to ensure data integrity. See, biology tolerates mutations/changes but software can be broken by changing 1 single bit. This requires redundant information storage and depends on later on sequencing practices. Anyhow, often readings are derived from a pool of viral particles, in which average sequence will be preserved. However, during human-to-human transmission, a bottleneck occurs. This determines the sequence of the newly formed pool and is called the founder effect. This interesting concept in population genetics tells that if a small group forms an isolated genetic pool (in this case few viral particles in a new host), the newly formed genetic pool may look different than the original genetic pool due to random sampling. This may break the computer code within the virus, without affecting the biological transmission. To counteract this, even more information storage is needed.
Another problem is the genome organization. If the viral genes and computer code are separated, this puts a selection pressure on the computer code, as it won’t be required for the biological transmission. Any deletion of the computer code part will favor this new form of the virus, as it can propagate much faster! An interleaved or even better overlapping genomic structure may preserve the computer code.
In contrast, computer transmission can much more easily preserve the biological code/genes. Transmission of kilobytes or even megabytes of data is rapid nowadays. Data integrity is ensured in the digital world. TCP/IP ensures with hashing and other techniques that the packet is sent accurately. Variations is the strength of biology. This ensures adaptation to new conditions. But biology is analog. Digital world doesn’t allow variation. Some computer viruses simulated this with polymorphism. This means that the code may change but the function is preserved. This isn’t exactly like mutations but it may help virus to avoid detection.
Overall, it is highly unlikely that such a virus can exist. Quality control methods in the pharma industry are very strict (GMP = Good Manufacturing Practice) and we anyhow don’t perform many gene therapies yet. Often, the sequence analysis is done on very large datasets, where almost any combination of short sequences occur by chance. Any bugs in the analysis code, would probably initially cause a crash early on. Operating systems are more secure and data handling can be performed on different machines. Even cloud helps to put another level isolation. From the biology side, size of the viral genome is a major restriction. An intact immune system probably will kill any virus, which isn’t already evolved to optimize human infection. It requires an amazing level of engineering to achieve a coding, which can work in both biology and computers. It would be interesting see bacteria, which contains both types of information. Nevertheless, transmission between different domains is extremely unlikely.
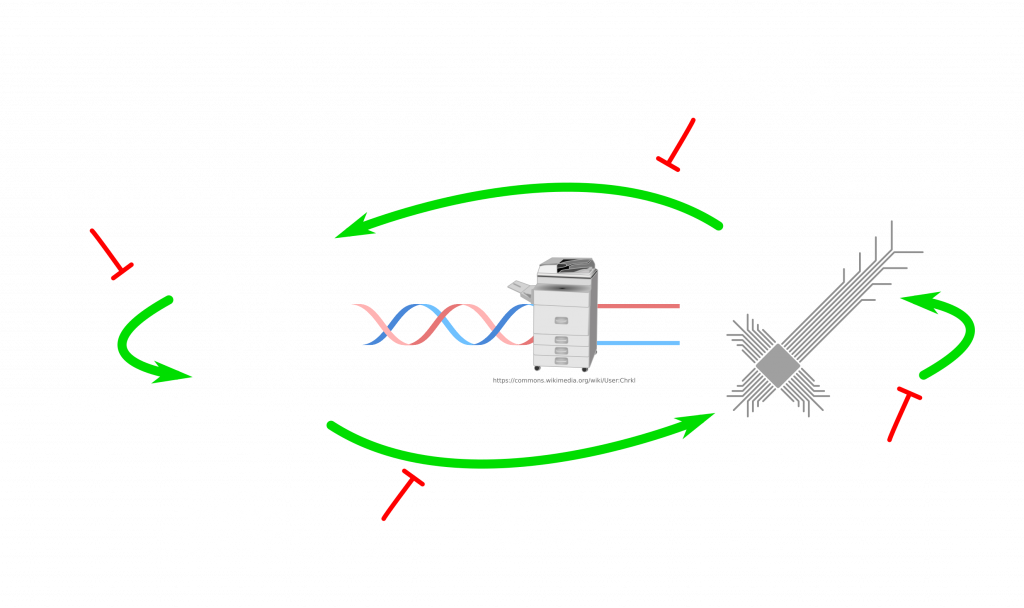
I guess, we will continue to have separate viruses for both domains. If anything like this ever tries, it would be more likely to be successful in a laboratory setting, where the life cycle is kept. Furthermore, bacteria and bacteriophages (viruses for bacteria) would be a better model. Deliberately introduced bugs can be exploited to prove the possibility. Both the synthesis and sequencing can be performed on the same controller computer for simplicity. Another interesting question is, if the biological evolution can help to improve the computer code of the virus? Maybe if the conditions are set right. If you think that this lifecycle is too complex to exist, look at this life cycle of a parasite:
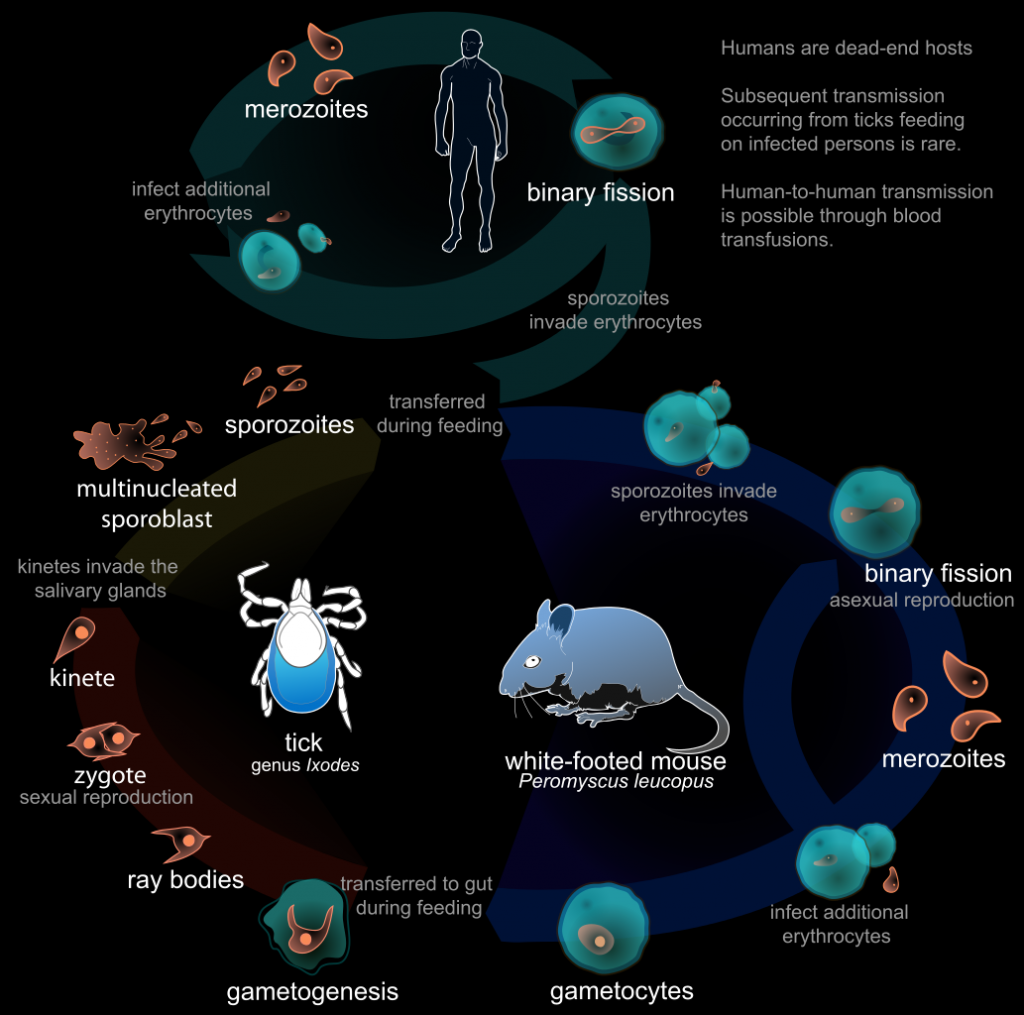
To study such a system, large containers with bacterial cultures with bacteriophages can be established. Over time, bacteriophages will kill some bacteria and some bacteria will evolve to evade. This culture must be kept stable, such that neither bacteria not the phages ever go extinct and it is a constant biological co-evolution.
Now let’s connect these containers with the previously mentioned sequencer/synthesizer systems. Introduce deliberate software bugs to be exploited! Reset systems in case of a fatal crash. Grab a popcorn and wait… I wonder if bacteriophages will manage to spread between systems. Perhaps, only infect one container with bacteriophages and keep the other containers free of them. Then see if biology can write a computer virus?!
I think, we should at least be aware of potential dangers of cyberattacks on pharmaceutical manufacturing plants. These could in the very best case stop the operations. In medical care, this can result in cost of life. Worst case scenario is that quality control mechanisms fail and batches of products can be contaminated. I doubt that the manufacturers are ignorant about this threat. This article shouldn’t scare anyone of seeking the help from modern medicine. It doesn’t scare me. Simply, I find the idea interesting and wanted to write about it. And it is longer then I have originally planned. If you like it, please feel free to share it.



